 Each atom in a molecule is identified by a unique serial number called the "atom id". Atom id's are automatically assigned by SYBYL when a molecule is created or input from a foreign source. SYBYL renumbers atom id's when a molecule description is modified.
Each atom in a molecule is identified by a unique serial number called the "atom id". Atom id's are automatically assigned by SYBYL when a molecule is created or input from a foreign source. SYBYL renumbers atom id's when a molecule description is modified.
 An alphanumeric tag, called the "atom name," is automatically assigned to each atom. Atom names are never changed automatically, but can be changed manually. Atom names can contain up to 7 characters from the set A-Z, "_", and "'".
An alphanumeric tag, called the "atom name," is automatically assigned to each atom. Atom names are never changed automatically, but can be changed manually. Atom names can contain up to 7 characters from the set A-Z, "_", and "'".
 An atom configuration code, called the "atom type," is assigned to each atom. Atom types specify the element and its hybridization (e.g. sp3 carbon, etc.). These codes are used by SYBYL to retrieve implicit atomic property information, such as van der Waals radii and force constants. Atom type codes are specified during the building process. Atom types specified in molecular structures input from non-SYBYL sources (e.g. CSD) are automatically translated to SYBYL types. This is not necessarily reliable, and should always be reviewed.
An atom configuration code, called the "atom type," is assigned to each atom. Atom types specify the element and its hybridization (e.g. sp3 carbon, etc.). These codes are used by SYBYL to retrieve implicit atomic property information, such as van der Waals radii and force constants. Atom type codes are specified during the building process. Atom types specified in molecular structures input from non-SYBYL sources (e.g. CSD) are automatically translated to SYBYL types. This is not necessarily reliable, and should always be reviewed.
The following atom type codes are currently available:
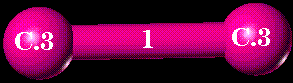
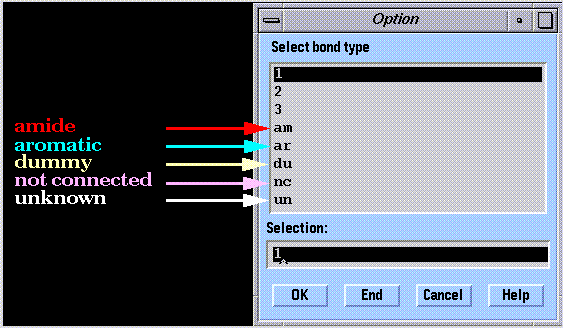
A bond order code, called the "bond type" is assigned to each bond in a molecule. Bond type and atom type codes are interdependent. The bond order is determined by the atom type codes of a pair of bonded atoms (conjugated systems can lead to ambiguities). Given the bond order and element names of a pair of bonded atoms, atom type assignments can be made by SYBYL. Again, this is not necessarily unambiguous.
The following bond types are available in SYBYL:

The rules can be reapplied as a structure is modified, allowing the dynamic set to be updated. The following rules are available:
Substructures are identified by a unique serial number called the "substructure id." The substructure organization allows particular monomers in a sequence to be specified without selecting their individual atoms. This can be used, for example, to set residue colors:
Special Considerations for Biopolymers
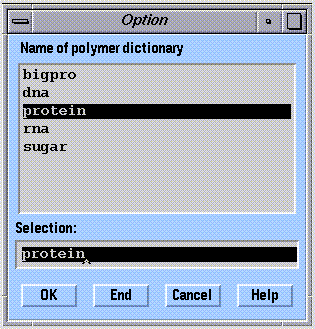
The molecule description input for biopolymers is handled somewhat differently than for other kinds of molecules. Biopolymers can be built from scratch using the "Biopolymer" option of SYBYL. The primary structure is specified from a list of monomer types, and the three-dimensional structure is automatically generated. The complete molecule description (monomer by monomer), in this case, is obtained from a "dictionary":
Several residues are distinguished by their degree and/or position of protonation. These choices affect the placement of protons by the "Add Hydrogens..." command of the "Biopolymer" menu. Pre-calculated charges are not available for use with the Tripos force-field, but charges can be calculated using the available methods in SYBYL. Pre-computed partial atomic charges are available for use with the Kollman force-field, and are input from the dictionary using the "Load Charges..." command of the "Biopolymer" menu.
The following non-standard amino acids are available:
The following N- and C-terminii configurations are available:
The following N- and C-terminal blocking groups are available:
Alternatively, biopolymer structures can be input from a PDB file. The dictionary is used, in this case, to assign everything but the atomic coordinates (which are taken from the PDB information). Some PDB files include a mixture of biopolymers and small molecules (e.g. a ligand). The small molecule description is automatically assigned by SYBYL, but often with poor accuracy (particularly atom type codes).
The Atom Expression
The atom expression contains a list of the atom id's of the atoms selected for graphical and computational operations. The atom expression can consist of a single atom or a collection of atoms that share a particular relationship (e.g. all in the same monomer). Atom expressions can be defined graphically or by command input. SYBYL provides a standard graphical interface for defining atom expressions:
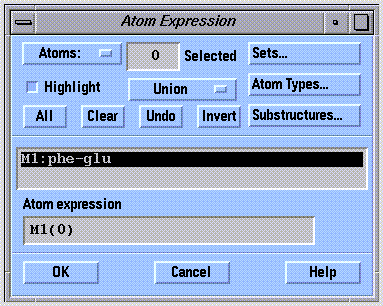
In monomer selection mode, the atom expression interface expects input at the monomer level. Clicking on an atom, for example, selects the entire monomer containing that atom. The monomer selection mode of the atom expression interface is shown below:
Alternatively, monomers can be selected from the text box, by dragging the mouse over the desired monomer id in the sequence:
Atom expressions can also be directly input from the keyboard or an SPL script. The following special characters are used to compose atom expressions using written commands:
The use of the above characters in the construction of atom expressions is illustrated in the following examples. Note that in many cases, there are multiple equivalent ways to construct atom expressions.
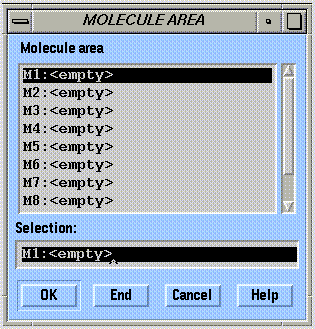
Select all atoms in a molecule (note that the molecule area need only be specified if more than one area is occupied):
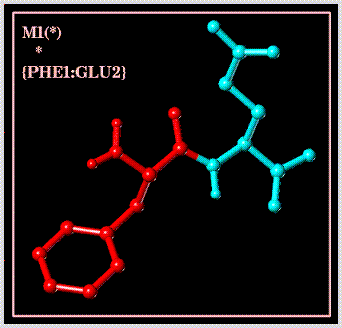
Select all atoms in a given monomer unit (substructure):
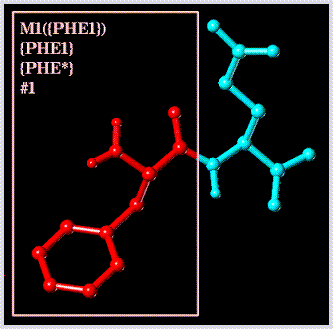
Select a particular atom by its "atom name" (note that the substructure containing the atom must be included in the atom expression because atom names can be identical in different substructures):
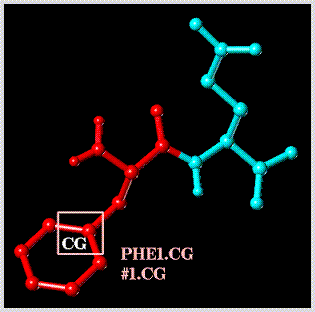
Select all atoms lying within a 2.5 Angstrom radius sphere centered about atom "CA" of "PHE1" (note that this involves the use of the "SPHERE" dynamic set rule):

Select all "C.AR" type atoms in substructure "PHE1":
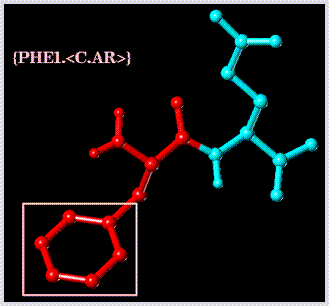
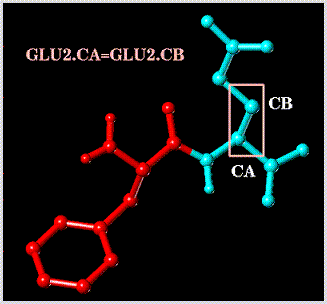
Select the bond defined by the dummy atoms named "CA" and "CB" of substructure "GLU2":